Engineered DNA nanotubes form tiny pipes into cells
06 Oct 2022 Simon Lichtinger
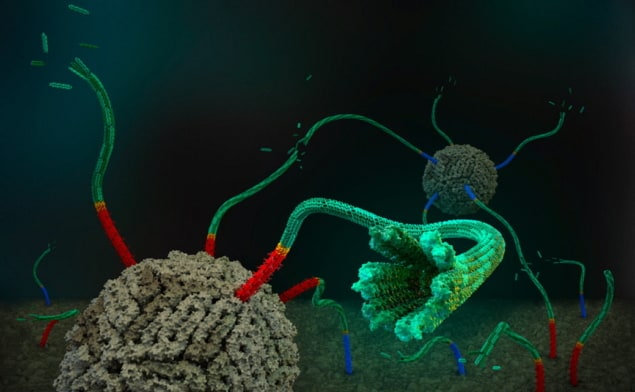
Nano plumbing: An artistic rendering showing how DNA nanotubes could connect artificial cells. (Courtesy: Schulman Lab, Johns Hopkins University)
Synthetic cells, engineered to mimic some of the functions performed by living cells, hold promise for applications in biotechnology and medicine. Even the smallest biological cells, however, are extremely complex and the construction of living artificial cells faces numerous roadblocks. Researchers in the Schulman Lab at Johns Hopkins University have recently made progress towards one of these challenges: the exchange of matter and information across cell boundaries.
Writing in Science Advances, the researchers – working in collaboration with the Aksimentiev Group at the University of Illinois Urbana-Champaign – demonstrate the leak-free transport of small molecules through engineered DNA nanochannels across unprecedented distances. In the future, their work may help in the construction of artificial cells, and also aid the study and manipulation of living tissue.
Cells within multicellular organisms need to exchange matter and communicate to ensure their collective survival. Since each cell is surrounded by a lipid membrane that is impenetrable to many biological molecules, evolution has produced mechanisms by which this barrier can be traversed. Signalling receptors, transporters and pores relay information and allow the passage of molecules between cells and their exterior, while cell contacts such as gap junctions directly connect the interior of neighbouring cells and enable cell-to-cell diffusion of small molecules.
To mimic these processes in artificial systems, “researchers have developed synthetic cells positioned next to each other that can communicate through protein pores on their membranes” explains first author Yi Li, who co-led the study. “However, developing synthetic cell systems where cells can communicate and exchange materials across longer distances is still a challenge.”
The protein structures that facilitate cell-to-cell communication in biology are built “bottom-up” from amino acids – the information encoded in their sequence translates into a structure. Another biological macromolecule, DNA, is mainly used for information storage in cells; but due to its ease of synthesis and potential to form high-level structures, the field of DNA nanotechnology has gone far beyond its first proof-of-concept some 30 years ago. Scientists have since assembled ever more sophisticated 2D and 3D structures from DNA, including lattices, tubes, geometric bodies and even artistic renderings of smiley faces, in efforts referred to as DNA origami.
In their study, the Schulman Lab researchers combined DNA origami nanopores, which bridge the membranes of cell-like vesicles and create small openings for molecules to cross, with engineered self-assembling DNA nanotubes. By quantifying the flux of a dye molecule into the vesicles, they showed that short nanopores made the membrane permeable to the dye. They also validated that the speed of this transport is consistent with diffusion and found that a specially designed DNA cap can block the pores and stop the dye from entering. First author: Yi Li in the laboratory at Johns Hopkins University. (Courtesy: Yi Li)
First author: Yi Li in the laboratory at Johns Hopkins University. (Courtesy: Yi Li)
 First author: Yi Li in the laboratory at Johns Hopkins University. (Courtesy: Yi Li)
First author: Yi Li in the laboratory at Johns Hopkins University. (Courtesy: Yi Li)The team then extended this work to DNA nanotubes with a median length of 700 nm and a maximum of over 2 µm. Again, experiments showed that dye influx is enhanced in the presence of the DNA constructs, and that the cap can arrest permeation. The implication, says Li, is that “small molecules can pass through the tubes without leaks, and we expect large molecules, such as proteins, can also be transported through these nanotubes”.
Members of the Aksimentiev Group conducted Brownian dynamics computer simulations of the nanopore–dye system. These illustrated that for molecules below a threshold size, leakage through the side wall of the DNA tube dominated influx, while for larger molecules, end-to-end diffusion becomes the preferred mechanism .
Li explains that such simulations are complementary with experiments in two ways. “They can be used as design tools to help researchers design nanoscale structures that have specific functions”, he says, for example by “simulating the self-assembly kinetics of our DNA nanostructures”, but they also help to “validate experimental results and provide additional insights into the physical processes”.READ MORE

Rebecca Schulman – who co-led the research – draws an analogy to pipes. “This study suggests very strongly that it’s feasible to build nanotubes that don’t leak using these easy techniques for self-assembly, where we mix molecules in a solution and just let them form the structure we want. In our case, we can also attach these tubes to different endpoints to form something like plumbing.”
The lab has ambitious plans for application of these nanotubes. “Future developments include connecting two or more artificial cells with our DNA nanotubes and showing molecular transport among them. We can potentially show [that] the transport of signalling molecules from one cell can activate/deactivate the gene expression in another cell,” Li tells Physics World. The team also hopes to “use nanotubes to control the delivery of signalling molecules or therapeutics to mammalian cells, either to study cell signalling behaviours or to develop a drug delivery strategy”.

Simon Lichtinger is a PhD student contributor to Physics World. Simon is at the University of Oxford, where he uses molecular simulations to study drug transport through membrane transporter proteins. Find out more about our student contributor networks
from physicsworld.com 2/11/2022
Δεν υπάρχουν σχόλια:
Δημοσίευση σχολίου